The US Government’s pursuit of the “Thousand Dollar Genome” and the “X Prize” goal of even lower cost DNA sequencing are striking examples of the importance being assigned to advances in methods for the DNA sequencing of the human genome. The first technology that finally allows for truly routine genome sequencing will open a new chapter in human medicine. It will revolutionize therapeutic clinical trials and start the realization of the promises of personalized medicine.
Three features will characterize the successful technology. First, it will need to perform “long reads.” Second, the successful technology will need to be scalable and suited for parallelism. Finally, the successful technology will need to operate on a single molecule basis. There have been many attempts to incorporate one or more of the above requirements into a practical DNA sequencing technology. Yet not one platform has surfaced that successfully incorporates the three features. Suggesting a fundamentally new approach to DNA sequencing is necessary.
Professor Stuart Lindsay at the Biodesign Institute of Arizona State University has developed a new a
The US Government’s pursuit of the "Thousand Dollar Genome" and the "X Prize" goal of even lower cost DNA sequencing are striking examples of the importance being assigned to advances in methods for the DNA sequencing of the human genome. The first technology that finally allows for truly routine genome sequencing will open a new chapter in human medicine. It will revolutionize therapeutic clinical trials and start the realization of the promises of personalized medicine.
Three features will characterize the successful technology. First, it will need to perform "long reads." Second, the successful technology will need to be scalable and suited for parallelism. Finally, the successful technology will need to operate on a single molecule basis. There have been many attempts to incorporate one or more of the above requirements into a practical DNA sequencing technology. Yet not one platform has surfaced that successfully incorporates the three features. Suggesting a fundamentally new approach to DNA sequencing is necessary.
Professor Stuart Lindsay at the Biodesign Institute of Arizona State University has developed a new approach to DNA sequencing. Lindsay’s technology utilizes single molecules. They are constrained and directed by nanoscale features and examined by a unique, DNA base-specific, electronic technique. Very long reads, many tens or even hundreds of thousands of bases, are enabled by this method.
The dimensions and design of this sequencer are such that plans are now in progress to fabricate large numbers of these on a silicon chip. Proofs of principle for the necessary electronics and software have been completed.
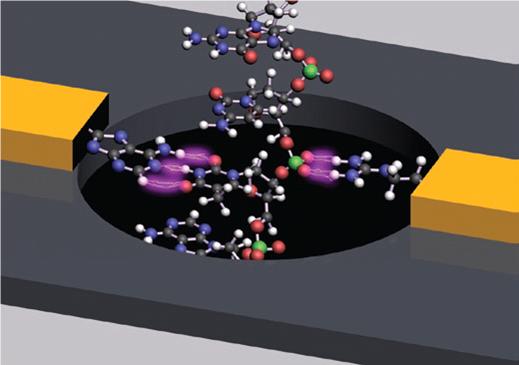
Approach to DNA sequencing. Lindsay’s technology utilizes single molecules. They are constrained and directed by nanoscale features and examined by a unique, DNA base-specific, electronic technique. Very long reads, many tens or even hundreds of thousands of bases, are enabled by this method.
The dimensions and design of this sequencer are such that plans are now in progress to fabricate large numbers of these on a silicon chip. Proofs of principle for the necessary electronics and software have been completed.
The Next Steps
The Biodesign Institute of Arizona State University, through its technology development and licensing company, Arizona Technology Enterprises (AzTE), is seeking investment partners to provide the capital for the final development, manufacturing scale up, and marketing of this technology. Interested parties will need to sign a confidentiality agreement to receive further information.